Introduction
R Statistical Language for EPR - A Comprehensive Rationale
Source:vignettes/introduction.Rmd
introduction.RmdThe usual way of a spectroscopist workflow consists in data transformation from the instrument via creation of figures and tables and finally using them in reports, publications, thesis or presentations. Quite often, several software packages/tools may be applied on this path, including the proprietary ones. Among them, an acquisition/processing software at the instrument enables basic or more advanced operations with the EPR outcomes/spectra. As already mentioned (see the README file / Homepage) there is an excellent open source toolbox for EPR specialists (1) only working under proprietary MATLAB (2). It can handle the raw files from the EPR machines process them and create reports in an interactive form (3) or in .pdf. There are, however, only a few examples of fully open source EPR packages, working under Python ![]() (4).
(4).
The open source R statistical language ![]() (5) is not so widely used as the general purpose language
(5) is not so widely used as the general purpose language ![]() , which is also applied in many scientific fields (6–8). Nonetheless, owing to its nature, the
, which is also applied in many scientific fields (6–8). Nonetheless, owing to its nature, the ![]() is very much focused on mathematics, statistics, graphing (it was actually built for such a purpose) and, last but not least, on data processing and analysis especially in research and academia. Even though so specialized, it actually represents an advantage in data handling. Therefore, it has been also very well established in chemistry and physics (9), biology (10), pharma industry (11, 12) as well as in finance area (13). Unsurprisingly, many big companies, universities or financial and healthcare institutions rely on a very robust and multiplatform1
is very much focused on mathematics, statistics, graphing (it was actually built for such a purpose) and, last but not least, on data processing and analysis especially in research and academia. Even though so specialized, it actually represents an advantage in data handling. Therefore, it has been also very well established in chemistry and physics (9), biology (10), pharma industry (11, 12) as well as in finance area (13). Unsurprisingly, many big companies, universities or financial and healthcare institutions rely on a very robust and multiplatform1 ![]() (14, 15).
(14, 15).
Because the ![]() belongs to the family of programming languages, it also provides reproducible workflow functionality. This is especially important for process validation in research and academia. Namely, one can easily follow all the data and visualization operations, unlike within the other software with “icon-based” procedures. Moreover, such a data wrangling and the related analytics, together with the outstanding visualization/graphing (16–19) and publishing capabilities (20–24), makes it a very powerful tool in the research area. Hence, the decision to create a package for EPR spectroscopy in
belongs to the family of programming languages, it also provides reproducible workflow functionality. This is especially important for process validation in research and academia. Namely, one can easily follow all the data and visualization operations, unlike within the other software with “icon-based” procedures. Moreover, such a data wrangling and the related analytics, together with the outstanding visualization/graphing (16–19) and publishing capabilities (20–24), makes it a very powerful tool in the research area. Hence, the decision to create a package for EPR spectroscopy in ![]() is obvious and strongly supported by the aforementioned features, even though the
is obvious and strongly supported by the aforementioned features, even though the ![]() is not so widely used in comparison to
is not so widely used in comparison to ![]() .
.
Moreover, the entire ![]() ecosystem with thousands of packages (5, 10, 25) and RStudio IDE2 (26) support workflows which are almost free from any other additional software/toolboxes (see also explanation below). Therefore, together with perfectly and uniformly structured documentation of R packages (which is already available within the RStudio) it represents an excellent facilitation of data processing workflows by extensive reduction of many steps which have to be otherwise performed by several programs, as already mentioned above. Great spectroscopy packages 📦 have been developed in
ecosystem with thousands of packages (5, 10, 25) and RStudio IDE2 (26) support workflows which are almost free from any other additional software/toolboxes (see also explanation below). Therefore, together with perfectly and uniformly structured documentation of R packages (which is already available within the RStudio) it represents an excellent facilitation of data processing workflows by extensive reduction of many steps which have to be otherwise performed by several programs, as already mentioned above. Great spectroscopy packages 📦 have been developed in ![]() e.g. for IR, MS, NMR, Fluorescence, UV-Vis(-NIR), Raman. However, non of them, even the general one like {ChemoSpec} or {hyperSpec}, are suitable for EPR, which actually possesses a special position among the spectroscopic techniques due to the paramagnetic (unpaired electronic state) nature of the studied molecules (27).
e.g. for IR, MS, NMR, Fluorescence, UV-Vis(-NIR), Raman. However, non of them, even the general one like {ChemoSpec} or {hyperSpec}, are suitable for EPR, which actually possesses a special position among the spectroscopic techniques due to the paramagnetic (unpaired electronic state) nature of the studied molecules (27).
Unlike the EasySpin (1), which is more concentrated on the simulation of EPR spectra, the primary aim of the eprscope package is to process, analyze and visualize the EPR spectral data similarly to functionality of the acquisition/processing software available at spectrometers. This is otherwise not available in any other software packages/toolboxes. Especially, the function performing an absolute quantitative EPR analysis is just a rarely present within the acquisition programs of the EPR instruments. Subsequently, the quantitative analysis is tightly connected with the determination of kinetic rate constants, \(k\) where the radicals (paramagnetic species) are involved in the studied chemical reactions. Last but not least, the \(k\) temperature dependence can be applied to determine the activation parameters (\(\Delta^{\ddagger} G^o\), \(\Delta^{\ddagger} H^o\) and \(\Delta^{\ddagger} S^o\) ) of the elementary radical reactions. Finally, the EPR spectroscopy is quite often coupled with the in situ (directly within the EPR cavity/probehead) radical formation techniques like electrochemistry (usually voltammetry or potentiostatic/galvanostatic electrolysis) and irradiation or UV-Vis(-NIR) spectroscopy (28). Therefore, the presented open source EPR ![]() 📦 will address not only the basic processing, simulation and visualization of the EPR spectra but also quantitative description of the radical reaction kinetics as well as that of the electrochemical redox ones. For the latter, this actually means that one could easily compare the number of transferred electrons from the voltammogram with the number of radical cations/anions determined by the quantitative EPR. Such information reveals if one-electron transfer is associated with the formation of a single radical, otherwise a more complex mechanism must be taken into account (28) (see also the
📦 will address not only the basic processing, simulation and visualization of the EPR spectra but also quantitative description of the radical reaction kinetics as well as that of the electrochemical redox ones. For the latter, this actually means that one could easily compare the number of transferred electrons from the voltammogram with the number of radical cations/anions determined by the quantitative EPR. Such information reveals if one-electron transfer is associated with the formation of a single radical, otherwise a more complex mechanism must be taken into account (28) (see also the eval_ECh_QNe_chronoamp() function).
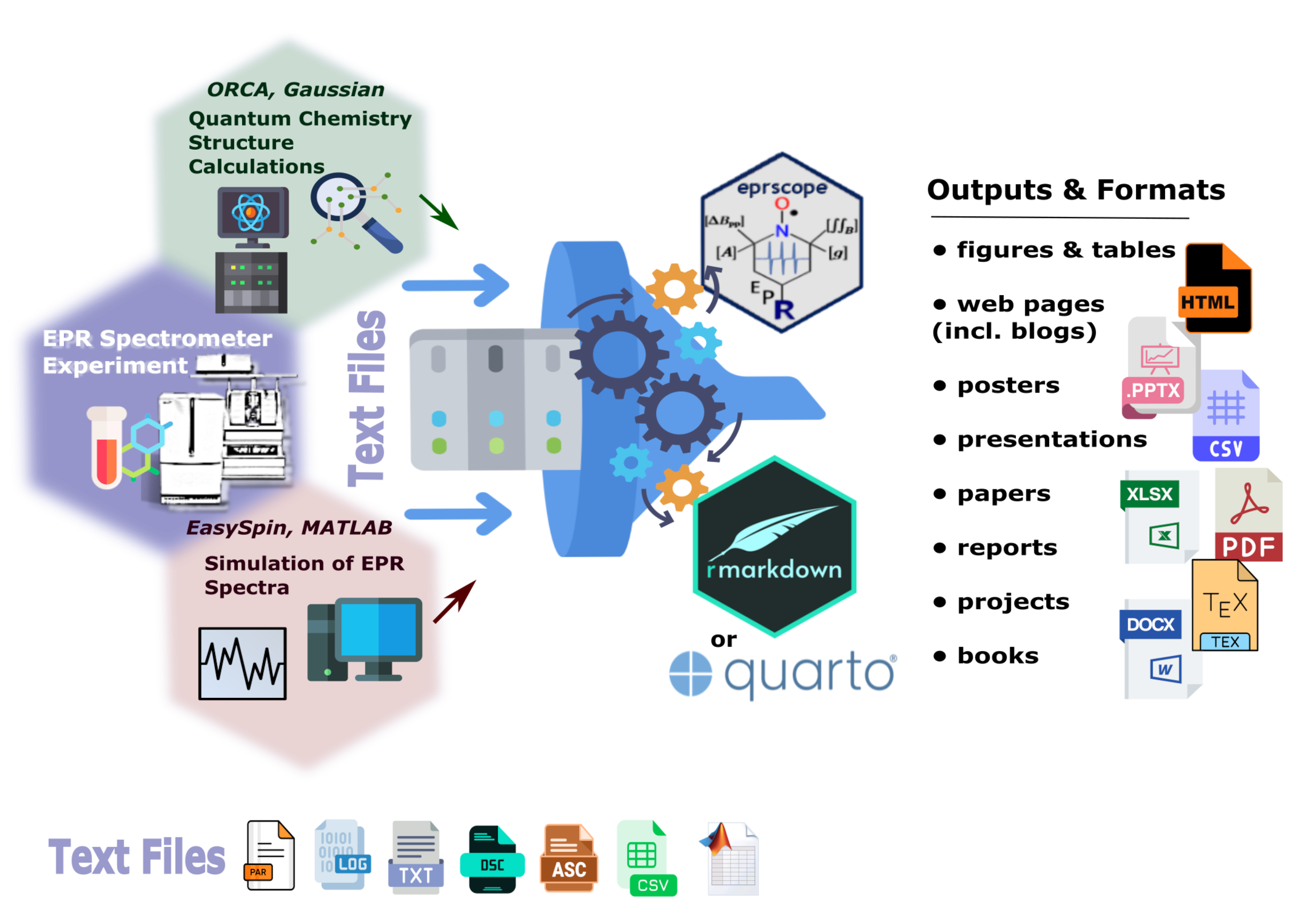
Figure 1: Schematic representation of the data workflow in EPR spectroscopy and the compatibility of the newly developed eprscope software package within the R ecosystem.
As mentioned above, the data analysis workflow also covers sharing of the results/outputs. However, quite often, in addition to essential processing of EPR spectra, the spectral analysis (which usually corresponds to determination of the hyperfine coupling/splitting constants and the g-factors) requires quantum chemical calculations on the high-performance computing servers and/or the above-mentioned simulations of EPR spectra done within the MATLAB by EasySpin3. Therefore, one must combine diverse outputs in order to gather the entire structural information about the paramagnetic center. As can be seen in Figure 1 the eprscope together with the Rmarkdown (23) and/or the Quarto publishing system (22) are capable (without living the ![]() ecosystem) to process all the input data into suitable dissemination form (e.g. report, presentation, manuscript, webpage…etc.) and thus very effectively completing the last stage of the reproducible research workflow (please, refer to the
ecosystem) to process all the input data into suitable dissemination form (e.g. report, presentation, manuscript, webpage…etc.) and thus very effectively completing the last stage of the reproducible research workflow (please, refer to the create_qmdReport_proj() function).